CNCPS Biology Model
Cornell Net Carbohydrate and Protein System (CNCPS) was developed and licensed by Cornell University Ruminant Nutrition Modeling Group. AMTS products use the latest version of CNCPS – version 6.5.5. By obtaining the license to the CNCPS model, AMTS can guarantee that the model is implemented exactly as developed.
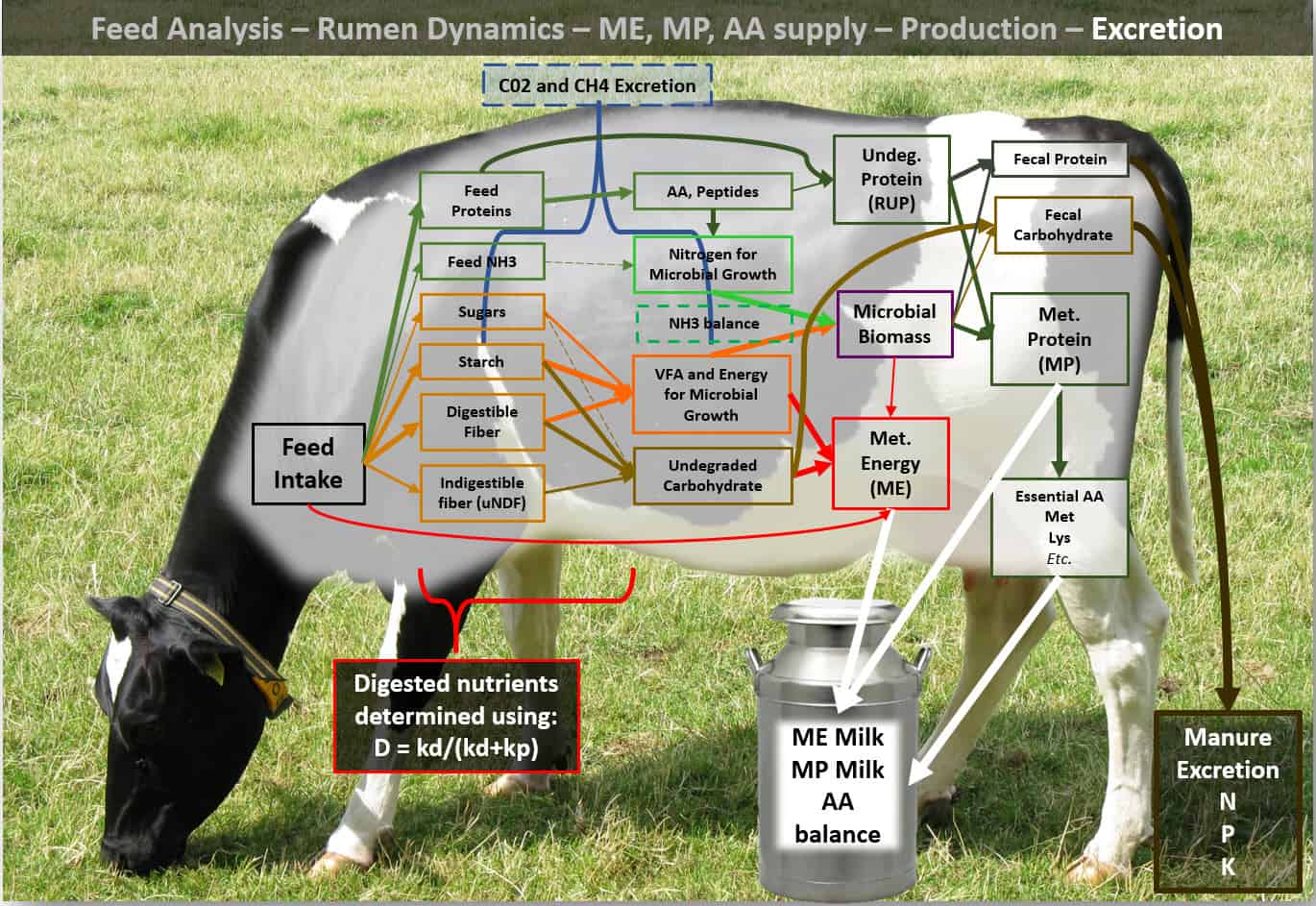
Models like the CNCPS are mechanistic models where nutrient utilization, primarily in the rumen, is described by a series of research derived, non-linear equations. Instead of having an NEL(Net Energy of Lactation) and a RUP (Rumen Undegradable Protein) value, feeds are described by their chemical fractions and fermentation characteristics. Then these “pools” are degraded (kd(digestion rate) / (kd + kp(passage rate))) and a microbial yield is predicted. Given differences in animal types, diets, etc., the total ME (Metabolizable Energy) and MP (Metabolizable Protein) is calculated for a diet. This is then partitioned to the animal requirements (maintenance, pregnancy, milk, growth, and reserves). What this allows us to do is more accurately define the diets. In today’s economic conditions and under today’s environmental scrutiny, this increased accuracy allows us to remove ration ‘safety factors’ that, in the past, led to nutrient overfeeding and greater environmental losses.
Proper use of the model has been shown via research to:
- improve cow health via
- decreased acidosis (clinical and subclinical)
- decreased laminitis and other foot problems
- decreased metabolic diseases incidence
- improve income over feed costs
- reduce nitrogen and phosphorus excretion
- improve animal performance (milk, meat, heifer growth, etc.)
- improve milk components
- place a higher farm awareness on forage quality

